Abstract
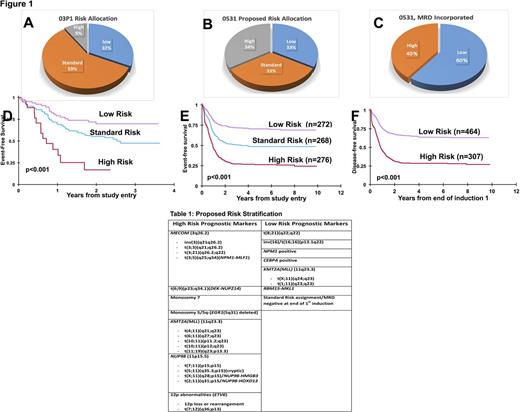
Introduction: Currently, most phenotypic, cytogenetic or molecular markers identified in children with acute myeloid leukemia (AML) are not used for risk stratification or treatment assignment. Historically, in the Children's Oncology Group (COG) pilot study AAML03P1, patients with core binding fusion (CBF) [t(8;21), inv(16)] or with NPM1 or CEBPA mutations had a more favorable outcome. Patients with FLT3 -ITD with high allelic ratio (FLT3 -ITD-HAR), monosomy 7, or -5/del(5q) had more adverse outcomes. In AAML03P1, 31% of patients were deemed low risk (LR), 9% were high risk (HR) and the remaining 60% were considered standard risk (SR) with event-free survival (EFS) of approximately 50% (Figure 1A). The same risk classification was carried forward to the successor Phase III COG study AAML0531 with similar outcomes. To develop an improved risk stratification strategy, we conducted a comprehensive retrospective evaluation of potential biomarkers that have been reported using available karyotype, immunophenotype and next generation sequencing (NGS) data from patients treated on AAML0531.
Methods: AAML0531 enrolled 1,022 patients (age 0-29 years) and randomized to standard therapy with or without gemtuzumab ozogamicin. Centrally reviewed data karyotype and FLT3, NPM1 and CEBPA status were available for this study. An additional 740 individual diagnostic specimens were interrogated by NGS (whole genome, transcriptome or targeted capture sequencing) to identify additional somatic variants including cryptic fusions, single nucleotide (SNV) and copy number variants (CNVs). Post induction response by multidimensional flow cytometry (MDF), an important prognostic modality, was also analyzed for risk stratification assignment in SR patients. Information from genomic and MDF parameters were analyzed for association with event free (EFS) and disease free survival (DFS).
Results: Table 1 shows the cytogenetic, molecular, and immunophenotypic markers used in our new risk stratification model. HR fusions detected by karyotype included 5 distinct KMT2A (MLL) fusion types, DEK-NUP214, NPM1 - MLF1, MECOM (3q26.2), and KAT6A (8p11.21). HR cryptic fusions detected by NGS include CBFA2T3-GLIS2, NUP98 family fusions (NUP98-NSD1, NUP98-KDM5A, NUP98-HMGB3 and NUP98-HOXD13) as well as ETS transcription factor family members (ETV6, ERG, FEV). NGS also identified HR MLL, t(6;9) or ETS fusions in 10% of patients who were initially reported as having a normal karyotype. HR stratification by immunophenotype analysis includes the RAM phenotype that was reported in younger patients with overlap with FAB-M7 as well as CBFA2T3-GLIS2 fusions.
When karyotype, immunophenotype, and NGS analyses were retrospectively applied to patients treated in AAML0531, the distribution of patients in each risk group changed significantly compared to AAML03P1 (Figure 1A and 1B). According to the revised risk stratification, HR variants constituted 34% of all AML patients and had poor prognosis (EFS-26.3% ± 5.4%). Patients with CBF AML, NPM1 or CEBPA mutations were included in the LR cohort and those with RBM15-MLK1 fusion were relocated to the LR cohort and these collectively constitute 33% of AML cases (EFS-70.4% ± 5.6%). The SR cohort was reduced to 33% in this proposed risk stratification schema (EFS-48.6% ± 6.2%). Minimal residual disease (MRD) by MDF at end of first induction was factored for SR patients in the proposed risk classification (Figure 1C) and found to have prognostic significance; therefore SR patients were assigned to either HR (DFS-28.5% ± 5.2%) or LR (DFS-63.7% ± 4.5%) cohorts.
Conclusions: Analysis of refined karyotyping and molecular data and incorporation of validated NGS data for patients in AAML0531 significantly altered the risk stratification used to predict relapse and assign patients to appropriate therapy. This report is the first to study a large number of patients assigned to a clinical trial and apply new standards for karyotyping, molecular and NGS. In future studies, we plan to study all patients on COG trials by NGS methods, validate additional novel biomarkers, and to identify new targets for future precision medicine-based clinical trials.
Loken: Hematologics Inc: Employment, Equity Ownership. Brodersen: Hematologics Inc: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal